Project P3
P3: MC-based Modelling, Monitoring, and Control of Microliter-scale Bioreactors
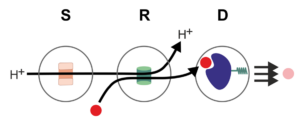
Motivation and state of the art:
The resource-efficient operation of microliter-scale bioreactors via the nanodevices developed in P1 requires accurate sensing and high precision control of information-carrying molecular signals. The modelling, sensing, and control of molecular signals1-3 and nanodevices4-7 can be investigated based on the MC paradigm. The resulting communication-theoretical models can be exploited, e.g., for the reduction of the number of resource consuming lab experiments needed for characterization of the nanodevices, the fine-tuning of the ratios of the protein components used for functionalization, the identification of scaling limits for decreasing system sizes, the design of the required communication and sensing processes, and the optimization of the overall control processes. In the literature, MC has been considered for the realization of synthetic communication systems4,8-11 and for the characterization of natural biological MC systems12-14. Several works have also investigated the controlled emission and detection of molecular signals via light-dependent proteins5. However, the MC concept has not been used in the context of microliter-scale bioreactors, yet. Therefore, this project will provide new tools for the modelling and control of bioprocesses and help initiate a new direction for MC research.
Objectives:
The goal of this project is the investigation of suitable models, signalling schemes, and algorithms for the optimal operation of the nanodevices developed in P1 for the monitoring and control of the reaction conditions in microliter-scale bioreactors. To this end, taking into account the involved chemical reactions and the geometry of the environment, the MC framework will be exploited to develop (i) analytical, numerical, and simulation models for the individual components of the pH-control, energy-converting, and substrate-feeding nanodevices investigated in P1 and P2, (ii) communication-theoretical models for all involved communication and sensing processes, and (iii) the required control signal designs and algorithms. The obtained models and resulting insights will also support the design and optimization of the nanodevices and protein modules studied in P1 and P2, respectively. For example, they can facilitate the optimization of the (average) number of protein modules to be inserted into a polymersome for optimal performance and minimal resource consumption.
References
1.Jamali, A. Ahmadzadeh, W. Wicke, A. Noel, and R. Schober. Channel Modeling for Diffusive Molecular Communication—A Tutorial Review, Proc. IEEE, vol. 107, pp. 1256-1301, Jul. 2019.
2.Suda, M. Moore, T. Nakano, R. Egashira, and A. Enomoto, “Exploratory Research on Molecular Communication between Nanomachines,” in Proc. Genetic Evol. Comput.. Conf., pp. 1-5, 2005.
3.Nakano, A. Eckford, T. Haraguchi. Molecular Communication, Cambridge University Press, 2012.
4.Söldner, E. Socher, V. Jamali, W. Wicke, A. Ahmadzadeh, H. Breitinger, A. Burkovski, K. Castiglione, R. Schober, and H. Sticht. “A Survey of Biological Building Blocks for Synthetic Molecular Communication Systems”. IEEE Commun. Surv. Tutor., vol. 22, pp. 2765-2800, Fourthquarter 2020.
5.Grebenstein, J. Kirchner, R. Stavracakis Peixoto, W. Zimmermann, F. Irmstorfer, W. Wicke, A. Ahmadzadeh, V. Jamali, G. Fischer, R. Weigel, A. Burkovski, R. Schober, “Biological Optical-to-chemical Signal Conversion Interface: A Small-scale Modulator for Molecular Communications,” IEEE Trans. Nanobiosci., vol. 18, pp. 31-42, Jan. 2018.
6.Brand, M. Garkisch, S. Lotter, M. Schäfer, K. Castiglione, and R. Schober, “Media Modulation in Molecular Communications”, in Proc. IEEE Int. Conf. Commun., Seoul, Republic of Korea, 2022, pp. 3697-3702, doi: 10.1109/ICC45855.2022.9838662.
7.Brand, M. Garkisch, S. Lotter, M. Schäfer, A. Burkovski, H. Sticht, K. Castiglione, and R. Schober, „Media Modulation Based Molecular Communication,“ IEEE Trans. Commun., vol. 70, no. 11, pp. 7207-7223, Nov. 2022.
8.Nakano, et al., “Molecular Communication Among Biological Nanomachines: A Layered Architecture and Research Issues,” IEEE Trans. Nanobiosci., vol. 13, pp. 169-197, Sep. 2014.
9.Bi, A. Almpanis, A. Noel, Y. Deng, and R. Schober, „A Survey of Molecular Communication in Cell Biology: Establishing a New Hierarchy for Interdisciplinary Applications,“ IEEE Commun. Surv. Tutor., vol. 23, no. 3, pp. 1494-1545, Thirdquarter 2021.
10.Yang, D. Bi, Y. Deng, R. Zhang, M. Ur Rahman, N. Ali, M. Imran, J. Jornet, Q. Abbasi, and A. Alomainy, “A Comprehensive Survey on Hybrid Communication in Context of Molecular Communication and Terahertz Communication for Body-centric Nanonetworks,” IEEE Trans. Mol. Biol. Multi-Scale Commun., vol. 6, pp. 107–133, Aug. 2020.
11.Kuscu and B. Unluturk, “Internet of Bio – Nano Things: a Review of Applications, Enabling Technologies and Key Challenges,” ITU J. Fut. Evolv. Technol., vol. 2, Dec. 2021, pp. 1-24.
12.Lotter, A. Ahmadzadeh, and R. Schober, „Synaptic Channel Modeling for DMC: Neurotransmitter Uptake and Spillover in the Tripartite Synapse,“ IEEE Trans. Commun., vol. 69, pp. 1462-1479, Mar. 2021.
13.Lotter, M. Schäfer, J. Zeitler, and R. Schober, „Saturating Receiver and Receptor Competition in Synaptic DMC: Deterministic and Statistical Signal Models,“ IEEE Trans. NanoBiosci., vol. 20, pp. 464-479, Oct. 2021
14.H. Arjmandi, D. Janzen, P. Gennemark, and A. Noel, “Do Cells Perceive Diffusion Noise?,” in Proc. 6th Workshop Mol. Commun., Coventry, UK, 2022.
